seaborn.pairplot¶
-
seaborn.pairplot(data, hue=None, hue_order=None, palette=None, vars=None, x_vars=None, y_vars=None, kind='scatter', diag_kind='auto', markers=None, height=2.5, aspect=1, dropna=True, plot_kws=None, diag_kws=None, grid_kws=None, size=None)¶ Plot pairwise relationships in a dataset.
By default, this function will create a grid of Axes such that each variable in
datawill by shared in the y-axis across a single row and in the x-axis across a single column. The diagonal Axes are treated differently, drawing a plot to show the univariate distribution of the data for the variable in that column.It is also possible to show a subset of variables or plot different variables on the rows and columns.
This is a high-level interface for
PairGridthat is intended to make it easy to draw a few common styles. You should usePairGriddirectly if you need more flexibility.Parameters: data : DataFrame
Tidy (long-form) dataframe where each column is a variable and each row is an observation.
hue : string (variable name), optional
Variable in
datato map plot aspects to different colors.hue_order : list of strings
Order for the levels of the hue variable in the palette
palette : dict or seaborn color palette
Set of colors for mapping the
huevariable. If a dict, keys should be values in thehuevariable.vars : list of variable names, optional
Variables within
datato use, otherwise use every column with a numeric datatype.{x, y}_vars : lists of variable names, optional
Variables within
datato use separately for the rows and columns of the figure; i.e. to make a non-square plot.kind : {‘scatter’, ‘reg’}, optional
Kind of plot for the non-identity relationships.
diag_kind : {‘auto’, ‘hist’, ‘kde’}, optional
Kind of plot for the diagonal subplots. The default depends on whether
"hue"is used or not.markers : single matplotlib marker code or list, optional
Either the marker to use for all datapoints or a list of markers with a length the same as the number of levels in the hue variable so that differently colored points will also have different scatterplot markers.
height : scalar, optional
Height (in inches) of each facet.
aspect : scalar, optional
Aspect * height gives the width (in inches) of each facet.
dropna : boolean, optional
Drop missing values from the data before plotting.
{plot, diag, grid}_kws : dicts, optional
Dictionaries of keyword arguments.
Returns: grid : PairGrid
Returns the underlying
PairGridinstance for further tweaking.See also
PairGrid- Subplot grid for more flexible plotting of pairwise relationships.
Examples
Draw scatterplots for joint relationships and histograms for univariate distributions:
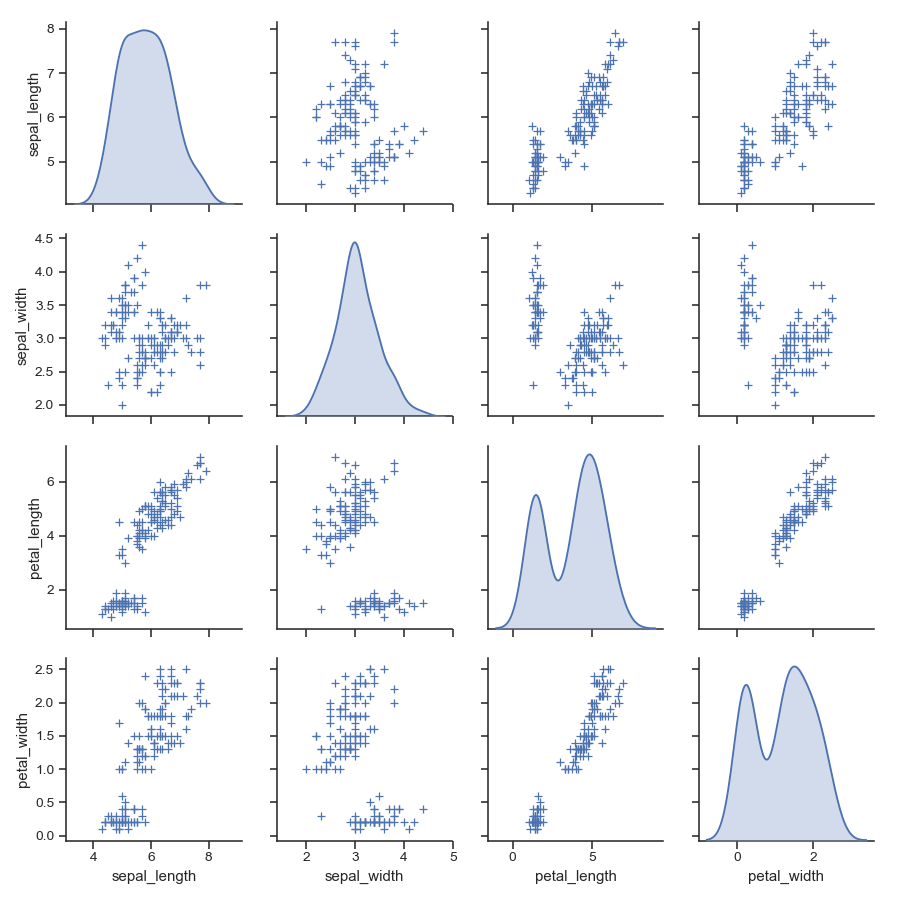
>>> import seaborn as sns; sns.set(style="ticks", color_codes=True) >>> iris = sns.load_dataset("iris") >>> g = sns.pairplot(iris)
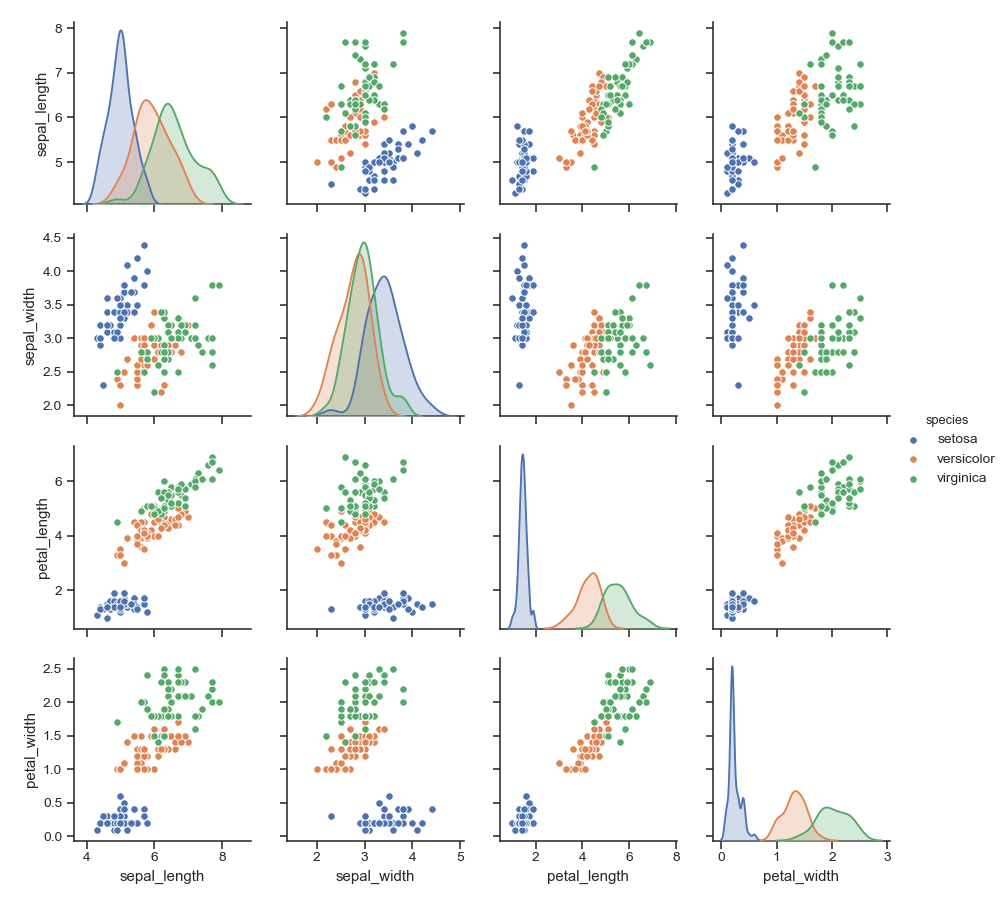
Show different levels of a categorical variable by the color of plot elements:
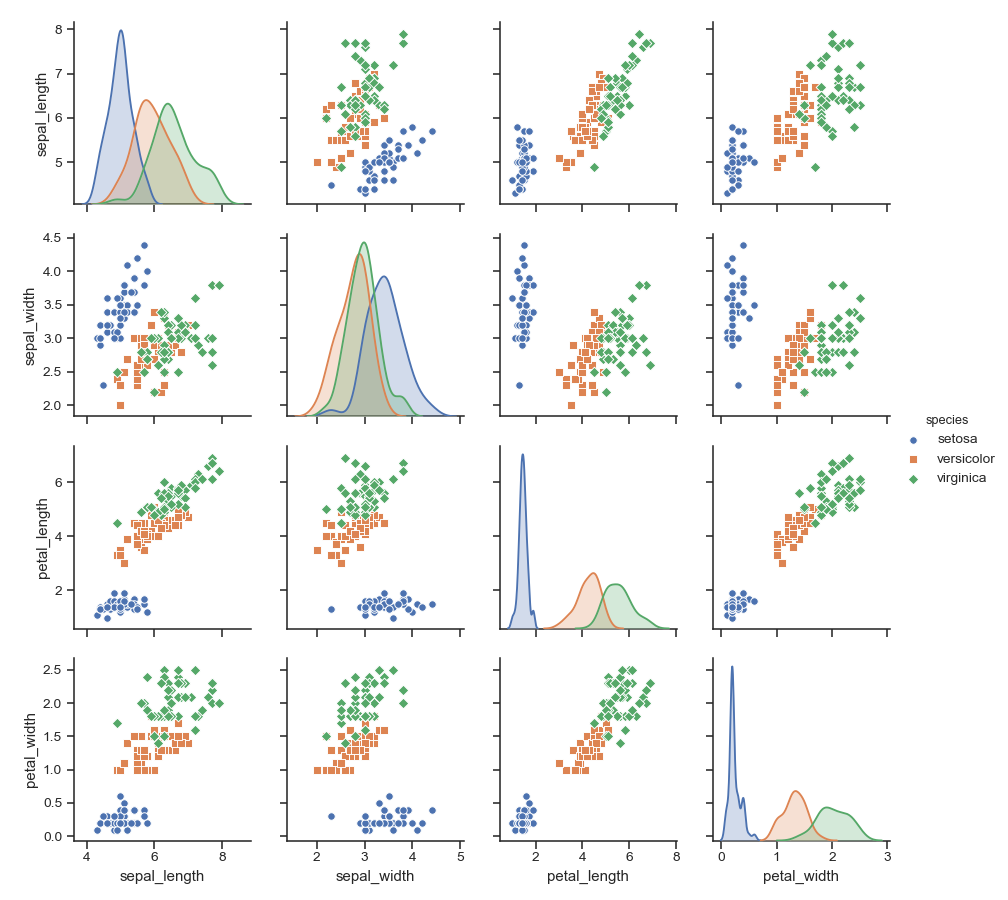
>>> g = sns.pairplot(iris, hue="species")
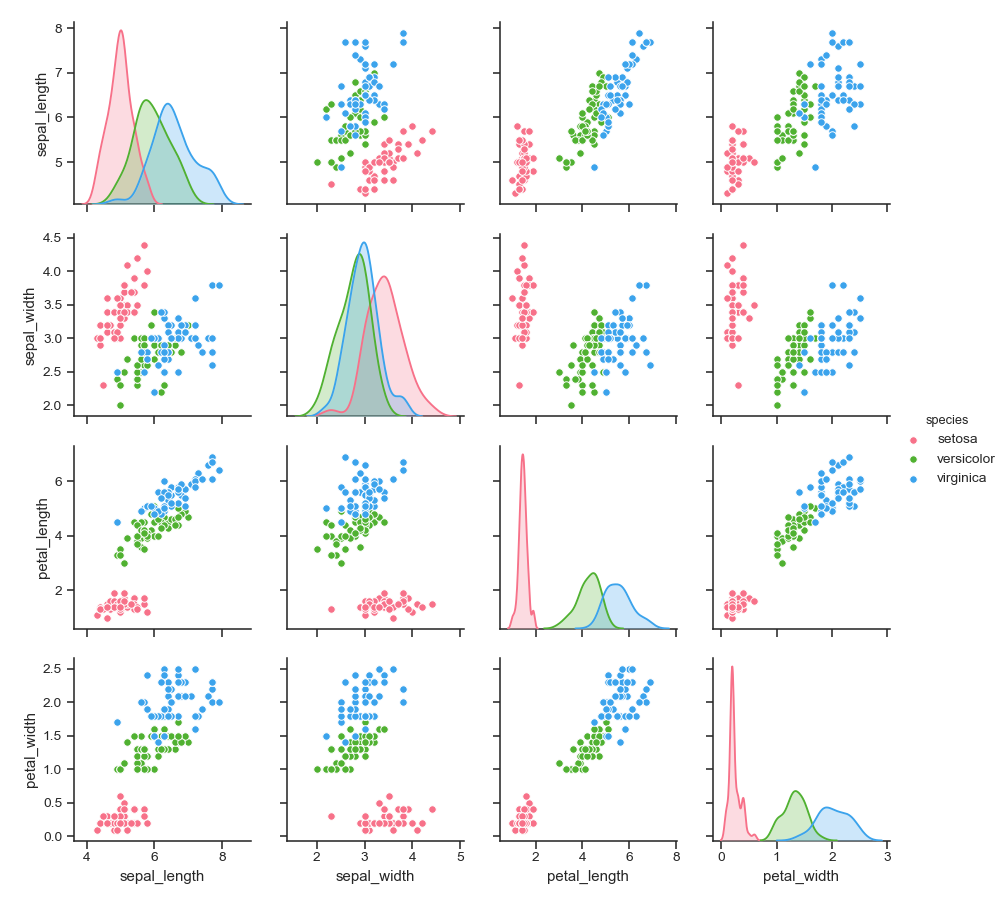
Use a different color palette:
>>> g = sns.pairplot(iris, hue="species", palette="husl")
Use different markers for each level of the hue variable:
>>> g = sns.pairplot(iris, hue="species", markers=["o", "s", "D"])
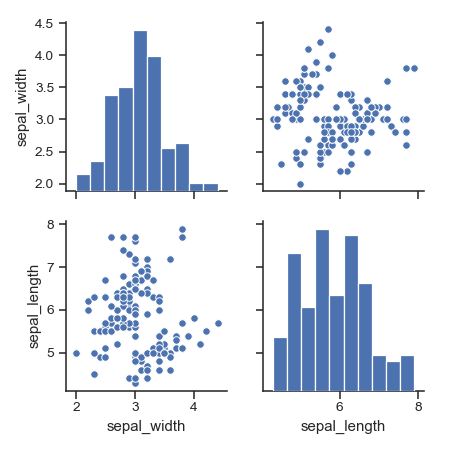
Plot a subset of variables:
>>> g = sns.pairplot(iris, vars=["sepal_width", "sepal_length"])
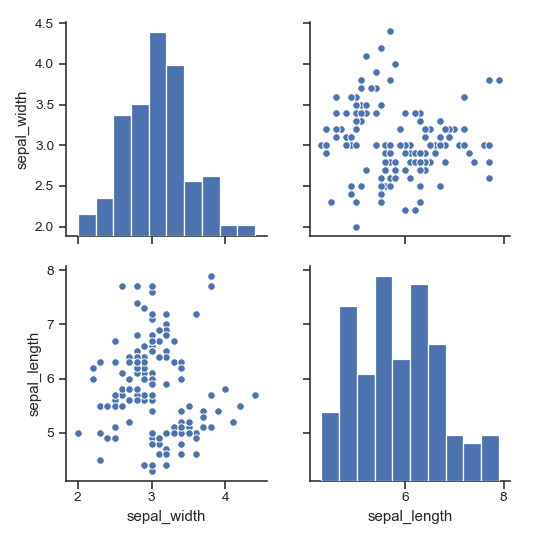
Draw larger plots:
>>> g = sns.pairplot(iris, height=3, ... vars=["sepal_width", "sepal_length"])
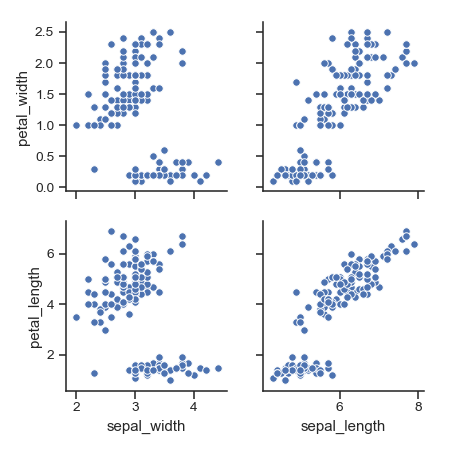
Plot different variables in the rows and columns:
>>> g = sns.pairplot(iris, ... x_vars=["sepal_width", "sepal_length"], ... y_vars=["petal_width", "petal_length"])
Use kernel density estimates for univariate plots:
>>> g = sns.pairplot(iris, diag_kind="kde")
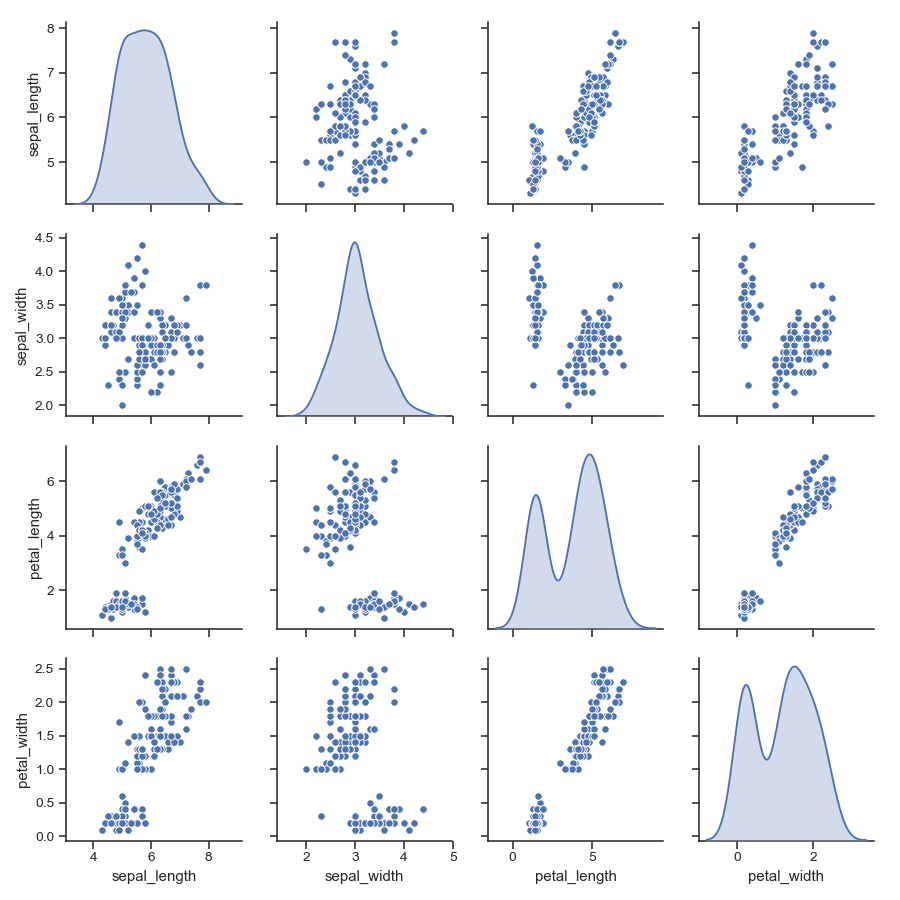
Fit linear regression models to the scatter plots:
>>> g = sns.pairplot(iris, kind="reg")
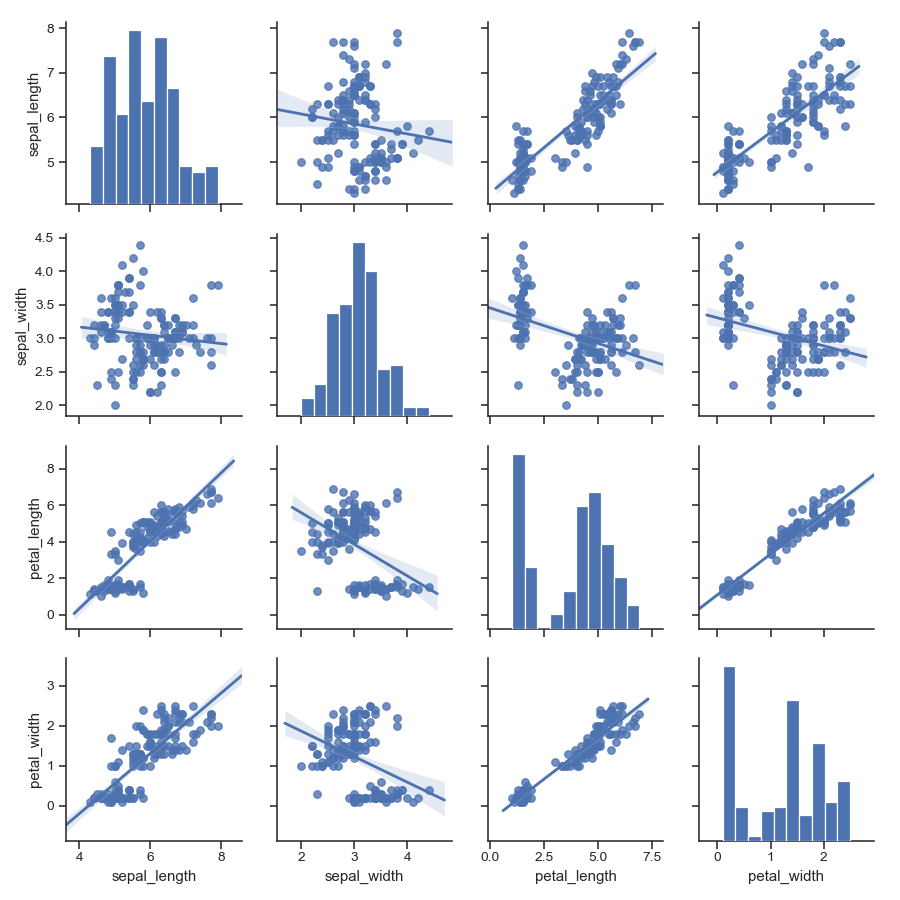
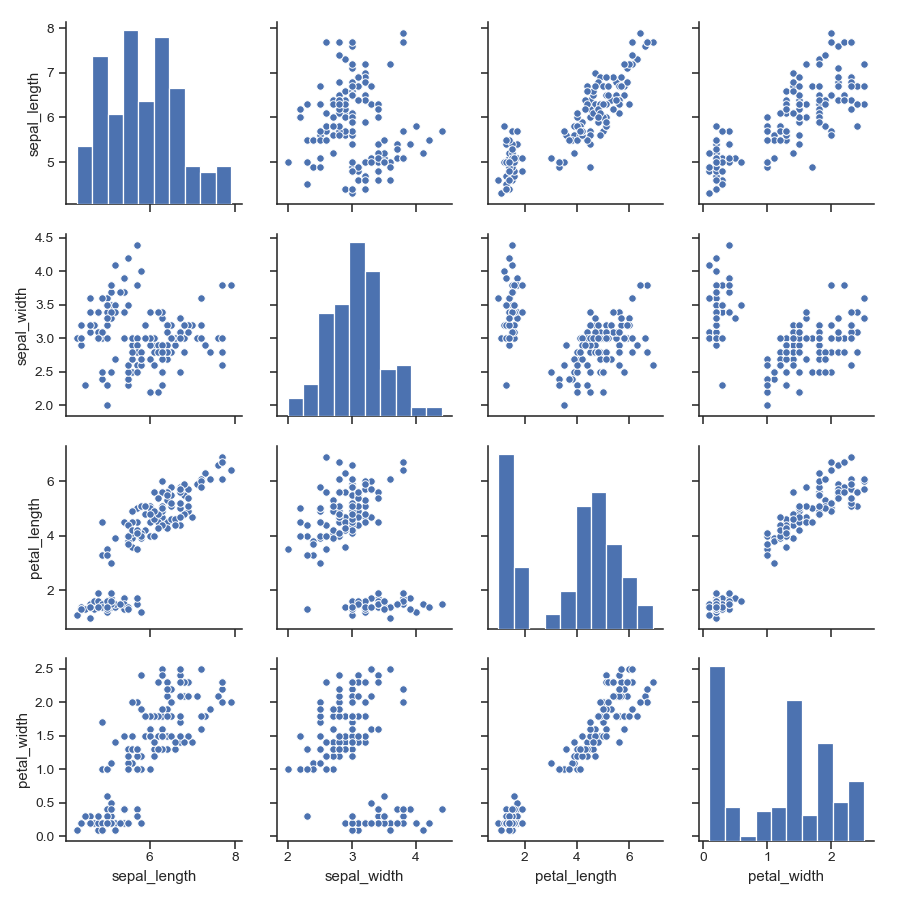
Pass keyword arguments down to the underlying functions (it may be easier to use
PairGriddirectly):>>> g = sns.pairplot(iris, diag_kind="kde", markers="+", ... plot_kws=dict(s=50, edgecolor="b", linewidth=1), ... diag_kws=dict(shade=True))