seaborn.heatmap¶
-
seaborn.heatmap(data, vmin=None, vmax=None, cmap=None, center=None, robust=False, annot=None, fmt='.2g', annot_kws=None, linewidths=0, linecolor='white', cbar=True, cbar_kws=None, cbar_ax=None, square=False, xticklabels='auto', yticklabels='auto', mask=None, ax=None, **kwargs)¶ Plot rectangular data as a color-encoded matrix.
This is an Axes-level function and will draw the heatmap into the currently-active Axes if none is provided to the
axargument. Part of this Axes space will be taken and used to plot a colormap, unlesscbaris False or a separate Axes is provided tocbar_ax.- Parameters
- datarectangular dataset
2D dataset that can be coerced into an ndarray. If a Pandas DataFrame is provided, the index/column information will be used to label the columns and rows.
- vmin, vmaxfloats, optional
Values to anchor the colormap, otherwise they are inferred from the data and other keyword arguments.
- cmapmatplotlib colormap name or object, or list of colors, optional
The mapping from data values to color space. If not provided, the default will depend on whether
centeris set.- centerfloat, optional
The value at which to center the colormap when plotting divergant data. Using this parameter will change the default
cmapif none is specified.- robustbool, optional
If True and
vminorvmaxare absent, the colormap range is computed with robust quantiles instead of the extreme values.- annotbool or rectangular dataset, optional
If True, write the data value in each cell. If an array-like with the same shape as
data, then use this to annotate the heatmap instead of the data. Note that DataFrames will match on position, not index.- fmtstring, optional
String formatting code to use when adding annotations.
- annot_kwsdict of key, value mappings, optional
Keyword arguments for
ax.textwhenannotis True.- linewidthsfloat, optional
Width of the lines that will divide each cell.
- linecolorcolor, optional
Color of the lines that will divide each cell.
- cbarboolean, optional
Whether to draw a colorbar.
- cbar_kwsdict of key, value mappings, optional
Keyword arguments for fig.colorbar.
- cbar_axmatplotlib Axes, optional
Axes in which to draw the colorbar, otherwise take space from the main Axes.
- squareboolean, optional
If True, set the Axes aspect to “equal” so each cell will be square-shaped.
- xticklabels, yticklabels“auto”, bool, list-like, or int, optional
If True, plot the column names of the dataframe. If False, don’t plot the column names. If list-like, plot these alternate labels as the xticklabels. If an integer, use the column names but plot only every n label. If “auto”, try to densely plot non-overlapping labels.
- maskboolean array or DataFrame, optional
If passed, data will not be shown in cells where
maskis True. Cells with missing values are automatically masked.- axmatplotlib Axes, optional
Axes in which to draw the plot, otherwise use the currently-active Axes.
- kwargsother keyword arguments
All other keyword arguments are passed to
matplotlib.axes.Axes.pcolormesh().
- Returns
- axmatplotlib Axes
Axes object with the heatmap.
See also
clustermapPlot a matrix using hierachical clustering to arrange the rows and columns.
Examples
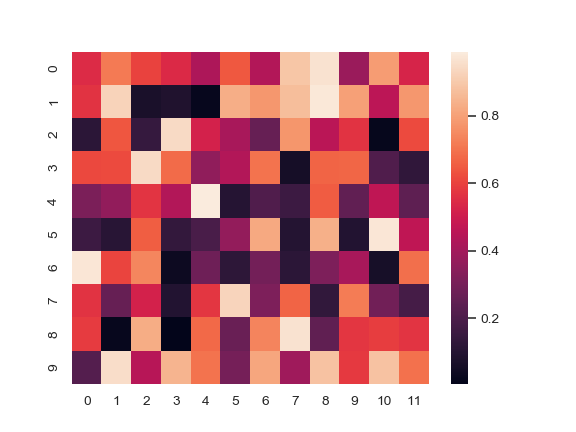
Plot a heatmap for a numpy array:
>>> import numpy as np; np.random.seed(0) >>> import seaborn as sns; sns.set() >>> uniform_data = np.random.rand(10, 12) >>> ax = sns.heatmap(uniform_data)
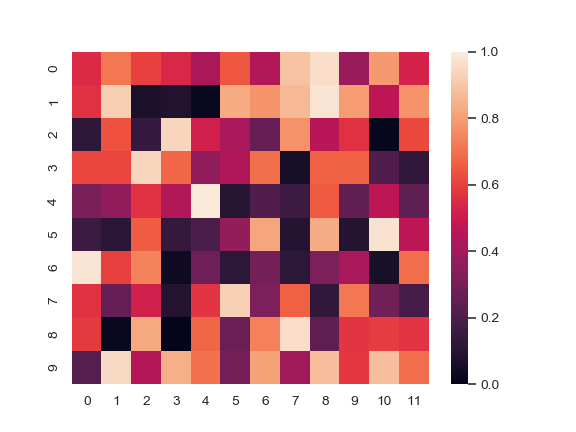
Change the limits of the colormap:
>>> ax = sns.heatmap(uniform_data, vmin=0, vmax=1)
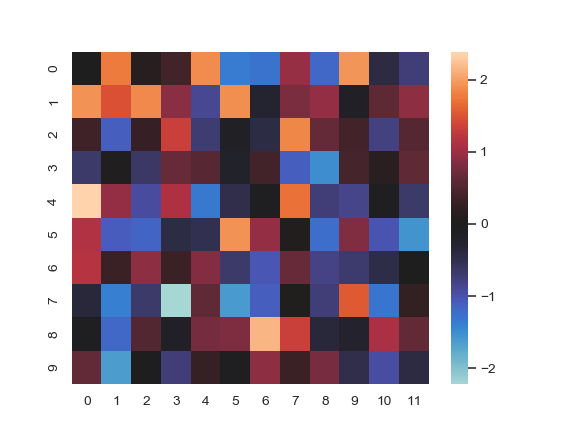
Plot a heatmap for data centered on 0 with a diverging colormap:
>>> normal_data = np.random.randn(10, 12) >>> ax = sns.heatmap(normal_data, center=0)
Plot a dataframe with meaningful row and column labels:
>>> flights = sns.load_dataset("flights") >>> flights = flights.pivot("month", "year", "passengers") >>> ax = sns.heatmap(flights)
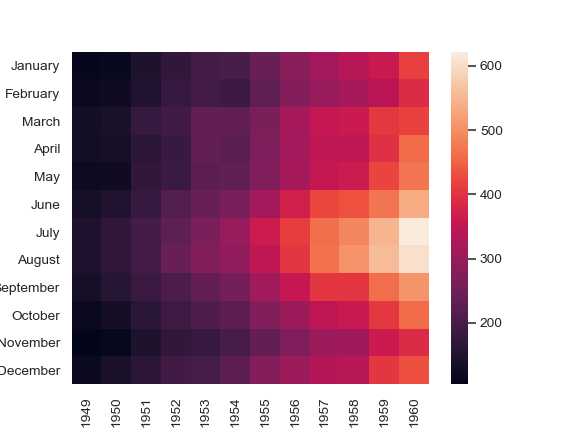
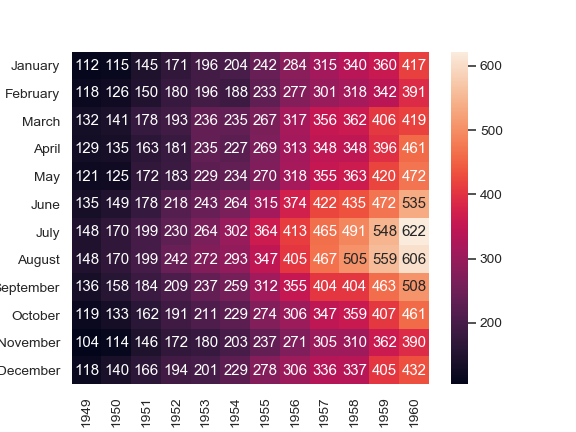
Annotate each cell with the numeric value using integer formatting:
>>> ax = sns.heatmap(flights, annot=True, fmt="d")
Add lines between each cell:
>>> ax = sns.heatmap(flights, linewidths=.5)
Use a different colormap:
>>> ax = sns.heatmap(flights, cmap="YlGnBu")
Center the colormap at a specific value:
>>> ax = sns.heatmap(flights, center=flights.loc["January", 1955])
Plot every other column label and don’t plot row labels:
>>> data = np.random.randn(50, 20) >>> ax = sns.heatmap(data, xticklabels=2, yticklabels=False)
Don’t draw a colorbar:
>>> ax = sns.heatmap(flights, cbar=False)
Use different axes for the colorbar:
>>> grid_kws = {"height_ratios": (.9, .05), "hspace": .3} >>> f, (ax, cbar_ax) = plt.subplots(2, gridspec_kw=grid_kws) >>> ax = sns.heatmap(flights, ax=ax, ... cbar_ax=cbar_ax, ... cbar_kws={"orientation": "horizontal"})
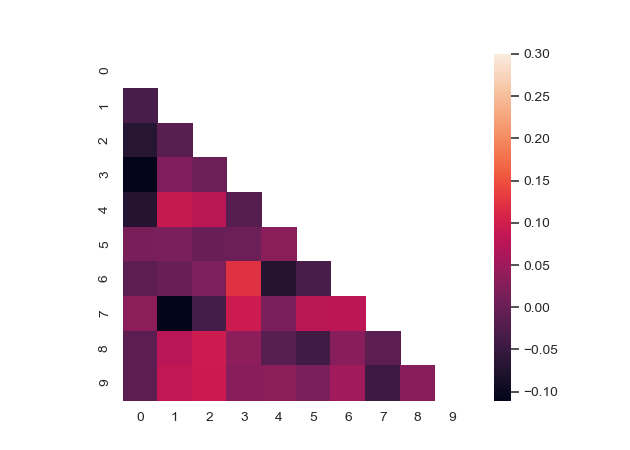
Use a mask to plot only part of a matrix
>>> corr = np.corrcoef(np.random.randn(10, 200)) >>> mask = np.zeros_like(corr) >>> mask[np.triu_indices_from(mask)] = True >>> with sns.axes_style("white"): ... f, ax = plt.subplots(figsize=(7, 5)) ... ax = sns.heatmap(corr, mask=mask, vmax=.3, square=True)